ENISI SDE: A new tool for modeling stochastic biological processes
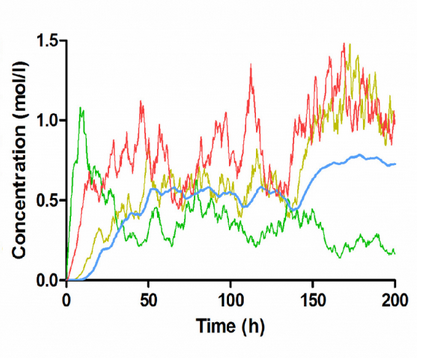
Stochasticity is part of many biological processes and an important aspect in modeling and simulations of computational biology. NIMML reseachers have developed a new user-friendly tool that integrated stochastic differential equations (SDE)-based modeling. The tool is available at here. A use case study has demonstrated the effectiveness of SDE as a stochastic modeling approach in modeling the intracellular signaling process underlying CD4+ T cell heterogeneity.
About NIMML
The NIMML Institute is a 501 (c) (3) non-profit public charity foundation focused on a transdisciplinary, team-science approach to precision medicine at the interface of immunology, inflammation, and metabolism. The NIMML Institute team has led numerous large-scale transdisciplinary projects and is dedicated to solving important societal problems by combining the expertise of immunologists, computational biologists, toxicologists, modelers, translational researchers, and molecular biologists. The Institute is headquartered in Blacksburg, VA. For more information, please visit www.nimml.org or contact pio@nimml.org.